The Genomics Team at the Medical Research Council Unit The Gambia at the London School of Hygiene & Tropical Medicine has successfully completed a study which generated Whole Genome data to determine the origin and pattern of transmission of SARS-CoV-2 from the first six cases tested in The Gambia.
The results of the study, published on bioRxiv shows that SARS-CoV-2 strains identified in The Gambia are of European and Asian origin, and the sequenced data matched the patients’ travel history. In addition, the study was able to demonstrate that the two COVID-19 positive cases travelling on the same flight had, in fact, different sources of infection.
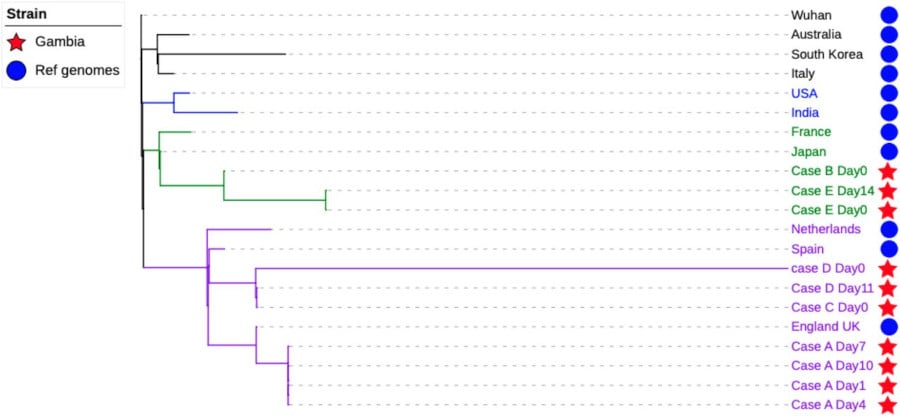
Nanopore and Illumina sequencers were successfully used to identify the sources of SARS-CoV-2 infection in COVID-19 cases in The Gambia. Sequencing reads were mapped to the Wuhan reference genome and compared to eleven other SARS-CoV-2 strains of Asian, European and American origins. A phylogenetic tree was constructed with the consensus genomes for local and non-African strains. Three of the Gambian strains had a European origin (UK and Spain), while two strains were of Asian origin (Japan).
All viruses mutate and none of the changes in SARS-CoV-2 has altered its behaviour or made it more dangerous, so far. The mutations could be as small as one of the roughly 30,000 biochemical letters that make up the virus. Like many viruses, SARS-CoV-2 stores its genes as RNA. Because the viral genome evolves at a steady rate as it replicates, averaging about 2.5 mutations a month, scientists can use this as a molecular clock. The genetic difference between SARS-CoV-2 samples gives an accurate estimate of when the lineages separated from one another.
Scientists studying mutations in coronavirus have decoded more than 10,000 different genomes of the deadly pathogen, creating a comprehensive map that will be crucial to controlling the pandemic and developing medicines to treat it.
International research teams have used phylogenetics to create a vast genealogical tree of the SARS-CoV-2 virus responsible for COVID-19, which reveals how it spread from the outbreak centre to all corners of the world.
Abdul Karim Sesay, Genomics Lab Manager at the MRCG at LSHTM said “We have successfully generated whole genome sequences of the first few cases of imported COVID-19 in The Gambia, in near real time during the current pandemic. This work is possible because of the collaboration between the MRCG at LSHTM and the Ministry of Health. Our data has provided a rapid genomics solution in the classification and identification of SARS-CoV-2 in The Gambia. I am very proud of my team, the MRCG at LSHTM staff, as well as our partners at the National Public Health Laboratories and the Ministry of Health”.
MRCG at LSHTM Director Prof. Umberto D’Alessandro said, “I am particularly pleased to see that our investments in the Unit’s genomics platform is proving to be very useful for the response to the COVID-19 epidemic. Sequencing the viral genome can provide precious information on the pathways of transmission that can be used to better target control efforts”.
With the Unit’s optimised and ready-to-go workflow, the Genomics team is set to generate data for tracking SARS-CoV-2 in The Gambia and other African countries within 24 hours of sample reception.
This will provide knowledge on the molecular epidemiology of COVID-19, give the true burden of the disease in this setting (as seen in the resolution of the indeterminate cases), provide information for African-specific vaccine development, and inform policy makers on decisions for strategic control measures.
Genomic epidemiology will be a vital tool in humanity’s efforts to beat COVID-19 and return to normal. It will be instrumental for helping to distinguish between local and imported transmissions if The Gambia moves out of the current state of emergency.
Our postgraduate taught courses provide health practitioners, clinicians, policy-makers, scientists and recent graduates with a world-class qualification in public and global health.
If you are coming to LSHTM to study a distance learning programme (PG Cert, PG Dip, MSc or individual modules) starting in 2024, you may be eligible for a 5% discount on your tuition fees.
These fee reduction schemes are available for a limited time only.