Malaria sporozoite detection during mosquito sugar feeding
Location of study: LSHTM and Guinea
LSHTM Investigators: Thomas Walker, Mojca Kristan, Victor Brugman
Funding Body: Royal Society
Rapid detection of infectious mosquitoes able to transmit malaria is of vital importance. Our aim was to determine if Plasmodium-infected Anopheles mosquitoes expel sporozoites during sugar feeding and if these sporozoites can be detected with sugar-coated FTA cards, providing a simple tool for sentinel site surveillance in malaria endemic settings. The ability to detect the presence of infectious mosquitoes is of vital importance for surveillance, control and elimination efforts, but can be labour-intensive and has to be carried out by trained personnel. In laboratory experiments we demonstrated that Anopheles mosquitoes infected with Plasmodium berghei and P. falciparum expel sporozoites during sugar feeding.
Sporozoite DNA was detected on sugar-soaked cotton wool and FTA cards using real-time PCR even when low numbers of sporozoites were ejected. These results demonstrate an effective and rapid methodology for detecting the presence of infectious mosquitoes with sporozoites and highlight potential laboratory applications for investigating mosquito-malaria interactions. Sugar feeding behaviour of adult female mosquitoes could also be exploited to detect malaria sporozoites in the wild, with FTA cards as a simple and economical tool used to enhance field surveillance activities for malaria. They preserve RNA and DNA without the need for a cold storage chain which is often problematic in tropical field settings. They were tested in a small field trial in Guinea, using modified CDC light traps but this was not effective due to low sporozoite rates. However, FTA cards have a potential to be used if further modified, allowing contact with larger numbers of infected mosquitoes over a longer period, not just a single night
Malaria vector biting behaviour
Location of study: Ghana and Guinea
LSHTM Investigators: James Orsborne, Thomas Walker, Laith Yakob
External collaborators: Yaw Afrane, (University of Accra, Ghana), Kalil Keita, (National Malaria Control Program, Guinea); Seth Irish, (CDC, USA)
Funding Body: MRC and Royal Society
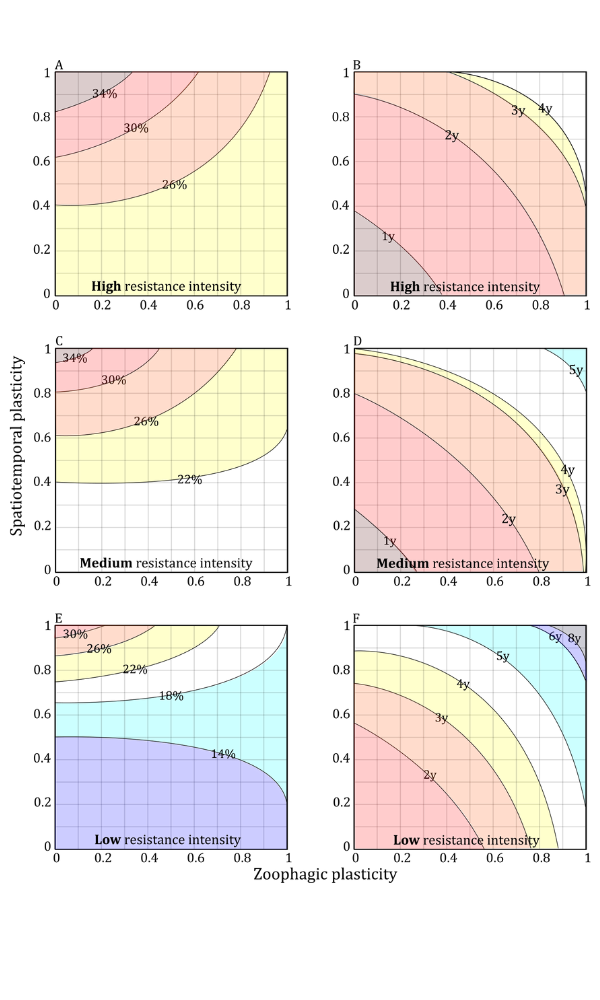
This research looks into how the mosquito vectors of malaria choose which host species to bite. Understanding how the environment can alter biting behaviour can inform how an environment could be altered to deflect bites away from people. It can also be used to inform targeting of different control tools.
A systematic review and meta-analysis of the human blood index of the major African malaria vectors revealed that the host choice was more associated with location of mosquito captures (indoors versus outdoors) than with mosquito (sibling) species, calling into question the dogma of ‘anthropophilic’ versus ‘zoophilic’ vector species. Using data-driven mathematical models, it was shown how understanding this biting behaviour is central to being able to project insecticide resistance frequency. It also underlies malaria control efficacy of bednets both in the short- and long-term.
Novel Wolbachia strains in Anopheles malaria vectors from Sub-Saharan Africa and their role in malaria transmission
Location of study: Guinea, Democratic Republic of the Congo (DRC), Ghana, Uganda , Madagascar
LSHTM Investigators: Thomas Walker, Claire Louise Jeffries, Mojca Kristan, James Orsborne, Kirstin Spence, Eliot Hurn
External collaborators: Gena G Lawrence and Seth R Irish, (Centers for Disease Control and Prevention, US); Janvier Bandibabone, (CRSN/LWIRO, DRC); Luciano M Tantely, Sébastien Boyer and Fara N Raharimalala, (Institut Pasteur de Madagascar, Madagascar); Kalil Keita, Denka Camara and Yaya Barry (Ministere de la Sante, Guinea); Francis Wat’senga and Emile Z Manzambi, (National Institute of Biomedical Research, DRC); Yaw A Afrane and Abdul R Mohammed, (University of Ghana, Ghana); Tarekegn A. Abeku, (Malaria Consortium, UK); Shivanand Hegde, Kamil Khanipov, Maria Pimenova and Yuriy Fofanov, George Golovko (University of Texas Medical Branch, US); Grant L Hughes (Liverpool School of Tropical Medicine, UK)
Funding Body: Wellcome Trust/Royal Society
Wolbachia are endosymbiotic bacteria found in ~40% of insects but historically thought to be absent in Anopheles. We discovered novel strains in species including Anopheles moucheti and we are currently investigating if high density strains can inhibit malaria transmission in Anopheles species and how they are maintained in mosquito populations.
We have analysed a range of Anopheles species to determine Wolbachia prevalence rates, characterise novel Wolbachia strains and determine any correlation between the presence of Plasmodium, Wolbachia, and the competing endosymbiotic bacterium Asaia. Anopheles adult mosquitoes were collected from five malaria endemic countries: Guinea, DRC, Ghana, Uganda and Madagascar, between 2013 and 2017. Molecular analysis has revealed a novel Wolbachia strains in species including An. coluzzii, An. gambiae s.s., An. arabiensis, An. melas, An. moucheti, An. species ‘A’ (A potentially new distinct species, as yet unnamed). Variable prevalence rates in different locations have been shown and novel strains are phylogenetically diverse. Wolbachia is the dominant member of the microbiome in An. moucheti and An. species ‘A’, but present at lower densities in An. coluzzii. The important discovery of novel Wolbachia strains in Anopheles provides greater insight into the prevalence of resident Wolbachia strains in diverse malaria vectors. Further experiments are planned to determine if high density Wolbachia strains inhibit malaria transmission and we will determine how these strains are being maintained in wild mosquito populations. Novel Wolbachia strains (particularly high density strains) are ideal candidate strains for transinfection to create stable infections in other Anopheles mosquito species, which could be used for population replacement or suppression control strategies.
Using human sera to profile antibody isotype responses to a panel of P. knowlesi-specific recombinant antigens.
Location of study: Sabah, Malaysia
LSHTM Investigators: Lou Herman, Tate Oulton, Katherine Glass, Kimberly Fornace, Chris Drakeley, Kevin Tetteh
External collaborators: Matthew Grigg, Nicholas Anstey, Timothy William Michael Blackman
Malaria cause by Plasmodium knowlesi is the most common form of the disease in Malaysian Borneo. It is a zoonotic disease transmitted from macaques to humans through mosquito bite. The true extent of the geographical boundaries for P. knowlesi transmission is currently unknown. The ability to measure antibodies to infection is a powerful tool which would help address this problem. We have developed a panel of recombinant proteins to be used as serological tools and to determine the antibody profiles this pathogen elicits. Antibody responses to Plasmodium spp. are known to play an important role in disease progression in infected hosts.
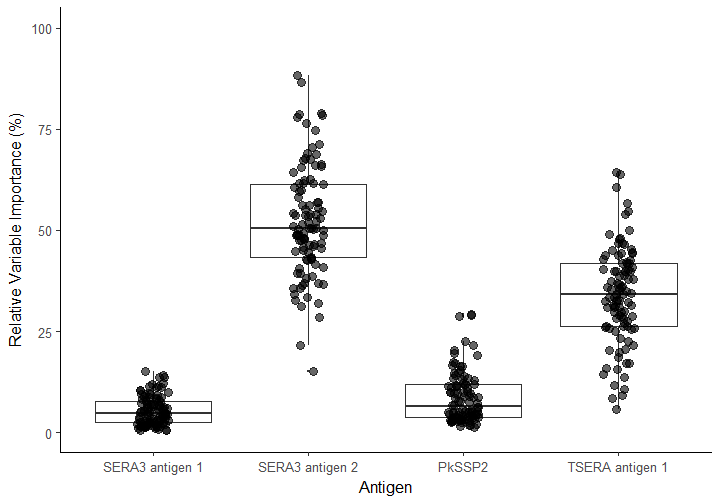
Much of the current data on antibody mediated protection and associated isotypes has come from human malaria but little is known about Plasmodium knowlesi. P. knowlesi is now the most common cause of malaria in Malaysian Borneo, with scarce data on the true extent of transmission and no data on the different reactivity profiles the disease elicits in endemic populations. A recently developed set of P. knowlesispecific antigens (n=10) were included in a protein microarray panel to determine antibody isotype (IgM, IgG and IgA) reactivity patterns for three post-treatment time-points from a hospital clinical treatment trial in Sabah, Malaysia (n = 12 individuals; 36 total samples for all time points) using labelled Qdot fluorescent probes. Strong reactivity of all isotypes to PkSERA3 antigen 2 was seen at all time points. IgG showed the strongest reactivity profile overall, followed by IgM which decreased over the course of follow up. IgA reactivity towards PkMSP1 antigen 2 and PKH_031930 antigen 2 mirrored that of IgM, indicating possible class switching from IgM to IgA for some antigens. These reagents represent novel tools with which to measure seroincidence to P. knowlesi. The development of such tools would help to answer questions relating to population exposure and our understanding of the geographical boundaries of infection. The differing isotype profiles in infections may be related to disease progression and severity – further work is need in this area.
Optimisation and Standardisation of a Reverse Phase Protein array (RPPA) for Malarial Serological Screening
Location of study: LSHTM
LSHTM Investigators: Tate Oulton, Katherine Glass, Will Stone, Kevin Tetteh
Funding Body: Global Good Fund - Intellectual Ventures
Serological screening for antibody responses in malaria is a proven approach for measuring exposure or investigating putative markers of protection, transmission and infectivity. The microarray platform is a high-throughput method for simultaneously detecting responses against hundreds to thousands of analytes, which we have rigorously optimised for our bespoke applications. We have evaluated materials, reagents and approaches to the manufacture and screening of protein microarrays for human IgG reactivity, including slide substrates, printing buffers, blocking buffers and a range of protein concentrations.
A core panel of purified proteins, of varying immunogenicity in reference samples, were printed and screened under 480 unique conditions and the performance under each condition compared.
A ratio of reactivity (Normalised Log2(Positive/Negative)) for each condition was calculated based on signals detected in the positive control versus the corresponding negative control, with conditions then ranked for each antigen. The top 10% of ranked conditions were compared between antigens to determine any commonalities, resulting in a list of eight conditions that performed best across the board. Interestingly, no condition performed consistently well for all antigens, with considerable heterogeneity in the reactivity ratio seen between antigens, within each condition. These data highlight the diverse behaviour of proteins in the context of a microarray; a multitude of variables such as size, charge, conformation and epitope availability likely have a considerable impact on performance under different assay conditions. Despite this, we have demonstrated that we are able to tailor a protein microarray platform that performs effectively, and will allow a substantial increase in the throughput of the serological assessment of malarial antigens as biomarkers for potential utility in disease surveillance.
Understanding the importance of the immunodominant CD8+ T cell epitope of Plasmodium circumsporozoite protein in parasite- and vaccine-induced protection
Location of study: LSHTM
LSHTM Investigators: Matthew Gibbins, Maya Glover, Jasmine Liu Julius Clemence Hafalla
External collaborators: Karolis Bauza, Arturo ReyesSandoval, (The Jenner Institute, Nuffield Department of Medicine, University of Oxford, UK); Katja Müller, Kai Matuschewski, (Humboldt University, Berlin, Germany); Olivier Silvie, (Sorbonne Université, Paris, France)
Funding Body: NC3Rs and The Royal Society
The circumsporozoite protein (CSP), the surface coat of sporozoites, is at the forefront of malaria vaccine development for the last 30 years. CSP has been shown to elicit strong CD8+ T cell responses that eliminate the developing parasites in hepatocytes resulting in protection. In this study, we characterised the importance of SYIPSAEKI, the immunodominant CSP-derived epitope of Plasmodium berghei in both sporozoite- and vaccineinduced protection in murine infection models.
In BALB/c mice, where SYIPSAEKI is efficiently presented in the context of the major histocompatibility complex class I (MHC-I) molecule H-2-Kd, we show that epitope-specific CD8+ T cell responses contribute to parasite killing following sporozoite immunisation. Yet, sterile protection is achieved in the absence of this epitope reinforcing that other antigens are vital for parasite-induced protective immunity. Furthermore, we establish that SYIPSAEKI-specific CD8+ T cell responses induced by viral-vectored CSP-expressing vaccines efficiently target parasites in hepatocytes and the resulting sterile protection strictly relies on the expression of SYIPSAEKI. We further show that in C57BL/6 mice, which expresses an irrelevant MHC-I and therefore unable to express the immunodominant epitope, CSP-based vaccines are unable to confer protection. These results further demonstrate the importance of CSP in protection against malaria preerythrocytic stages and that a significant proportion of the protection against the parasite is mediated by CD8+ T cells that are specific for the immunodominant epitope of this sporozoite surface protein.
India ICEMR – Pathogenesis of Cerebral Malaria in India
Location of study: Ispat General Hospital, Rourkela, India, LSHTM
LSHTM Investigators: Sam Wassmer
External collaborators: Jane Carlton & Steven Sullivan (New York University, US); Sanjib Mohanty, (Centre for the Study of Complex Malaria, India); Rajyabardhan Pattnaik, Megharay Majhi & Rashmi Ranjan Mohanty (Ispat General Hospital, India); Praveen Sahu, (Centre for the Study of Complex Malaria, India); Akshaya Mohanty, (Institute of Life Sciences, India); Angelika Hoffmann, (Heidelberg University Hospital, Germany); Joe Smith, (Center for Infectious Disease Research, USA)
Funding Body: National Institutes of Health (NIH), USA
This project is part of an International Center of Excellence for Malaria Research (ICEMR) programme, which was created in July 2010 by the NIH. The aim of the programme is to establish a global network of independent research centres in malaria-endemic countries to provide knowledge, tools, and evidencebased strategies to support malaria researchers working in a variety of settings.
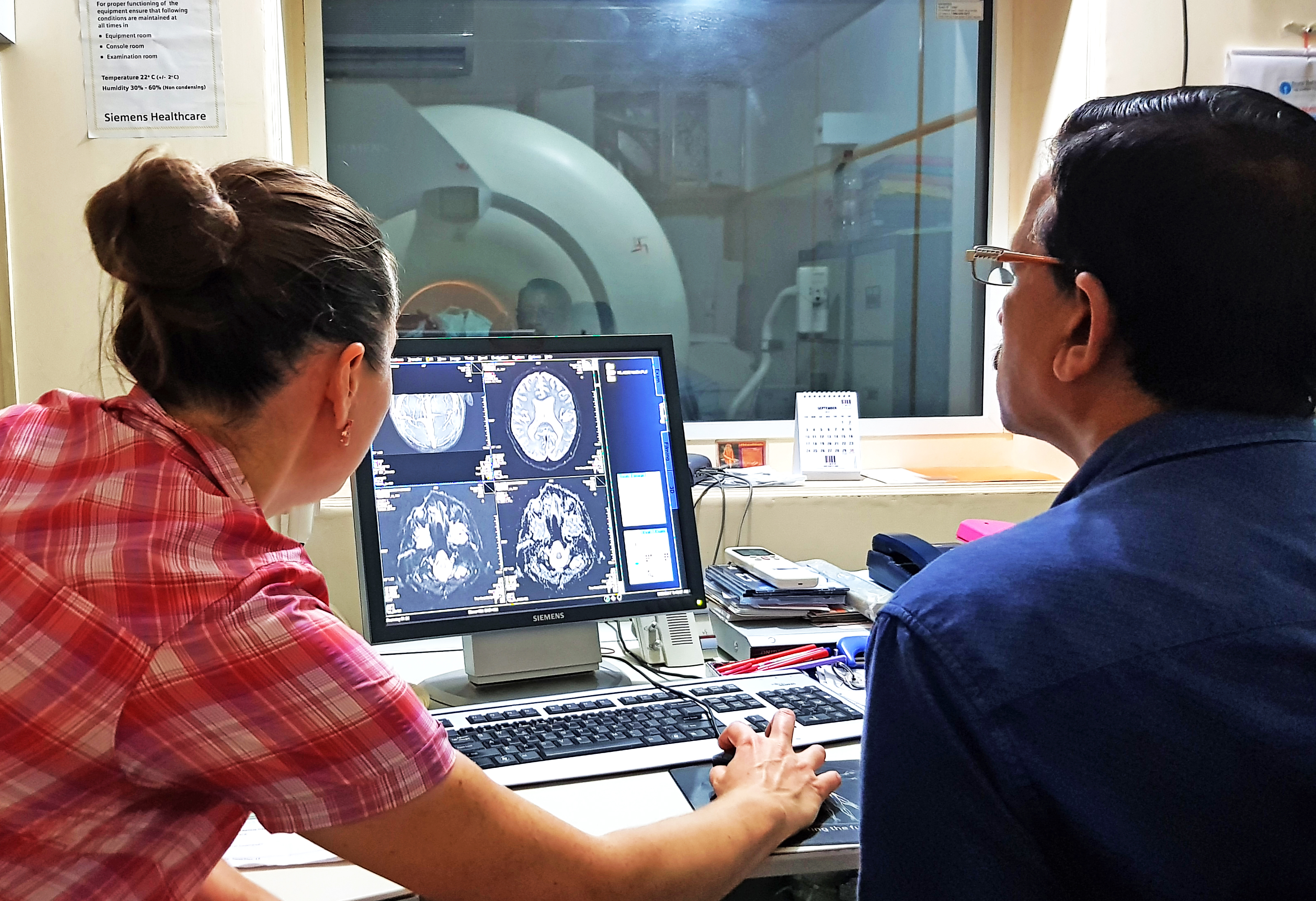
Plasmodium falciparum malaria is a major cause of mortality and morbidity in the developing world and cerebral malaria (CM), its most severe form, accounts for the majority of malaria-associated deaths. The pathophysiology and the molecular mechanisms underlying this complex neurologic syndrome are still poorly understood. Our team is using a unique combination of advanced neuroimaging techniques, post-mortem analyses, molecular biology approaches and machine-learning to explore the origin of brain swelling in CM patients from India. In a recent study carried out at Ispat General Hospital in Rourkela, we showed for the first time that increased brain volume during CM is predominantly due to posterior vasogenic oedema, indicating a local impairment of the bloodbrain barrier integrity. In half of the scanned patients, we found that blood vessel congestion in the basal ganglia also contributed to the swelling, a phenomenon likely caused by a considerable accumulation of P falciparum-infected red blood cells. Our team is now looking into potential associations between specific PfEMP-1 variants in the blood of CM patients and the presence or absence of vasogenic oedema identified by MRI. We are also applying advanced machine-learning models of CM disease causation, informed by clinical and laboratory investigations. The primary outcome of this project will be a better understanding of the different pathogenetic processes involved in CM, which will guide the development of new adjunct therapie
Unravelling the immune signature of Plasmodium falciparum transmission reducing immunity
Location of study: UK, Netherlands, Burkina Faso, The Gambia, Cameroon
LSHTM Investigators: Will J. R. Stone, Colin J. Sutherland, Sophie Jones, John Bradley, Chris Drakeley
External collaborators: xx
Funding Body: Marie Curie Career Integration Grant
When people are infected with malaria parasites they develop antibodies. These help kill the parasites, and in rare cases also stop mosquitoes becoming infected when they take a blood-meal. We investigated this natural transmission blocking immunity, identifying antibody responses correlated with the phenomenon that could form the basis of a new transmission-blocking malaria vaccine.
Plasmodium infection can elicit antibodies that inhibit the parasites survival in the mosquito, when they are ingested in an infectious blood meal. We determined the transmission-reducing activity (TRA) of naturally acquired antibodies from more than 600 malaria infected individuals in lab based mosquito-feeding assays. Transmission inhibition was associated with antibody responses to the Plasmodium proteins Pfs48/45 and Pfs230, as well as 43 novel gametocyte proteins. In field-based transmission assays, mosquito infection was significantly lower when they fed on the blood of individuals with antibodies specific to Pfs48/45, Pfs230, or to combinations of the novel TRA associated proteins. We also showed that naturally acquired purified antibodies against Pfs48/45 and Pfs230 are mechanistically involved in TRA (i.e. they can block mosquito infection alone), while human sera depleted of these antibodies retained high-level, complement independent TRA. Together, our analysis indicates that humans naturally develop as yet uncharacterised transmission blocking antibodies in response to malaria infection, and that antibody responses to novel gametocyte proteins are associated with reduced malaria transmission efficiency from humans to mosquitoes. A number of these proteins are now being assessed as potential transmissionblocking vaccine candidates.
Serological profile of antibody responses to P. falciparum in Guapi, Columbia
Location of study: LSHTM, the Wellcome Trust Sanger Insitute (WTSI) and Universidad Nacional de Colombia in Bogotá (UNC)
LSHTM Investigators: Tate Oulton, Katherine Glass, Chris Drakeley and Kevin K. A. Tetteh
External collaborators: Angelica Knudson, Alena Pance, Julian Rayner, and Vladimir Corredor (Universidad Nacional de Colombia in Bogotá (UNC), Wellcome Trust Sanger Institute (WTSI))
The Guapi region, on the Pacific Coast of Columbia, is emerging from decades of armed conflict, forced displacement and a lack of basic healthcare infrastructure. Understanding the disease landscape after such a protracted period of conflict will be essential to addressing public health needs in the region. A fifth of the malaria cases in South America occur in Columbia and of this, ~70% occur in the Pacific coast region.
The ongoing peace process has allowed a window of opportunity to access and serologically profile malaria responses in the area. Guapi, a town and municipality in the Cauca department of Columbia, lies within this region. The population is of largely African decent, and therefore resistant to P.vivax infections. As such, malaria in this region is predominantly caused by Plasmodium falciparum. Using a recently developed recombinant protein microarray, we have assayed serum samples from 1373 volunteers in Guapi, to obtain a serological profile of responses to a panel of >120 recombinant proteins, based on P. falciparum. The antigens were expressed in either E. coli or mammalian expression cells (HEK293E). The most recognised antigens included GLURP R2, AMA1, a variant of MSP2 (15-38) and a conserved membrane antigen of unknown function (Pf3D7_1419400). Work is ongoing to fully characterise antibody responses in the region, complimenting additional work focused on describing the factors unique to the locality that may act as barriers to malaria elimination.
Global genetic diversity of var2csa in Plasmodium falciparum with implications for malaria in pregnancy and vaccine development
Location of study: LSHTM
LSHTM Investigators: Ernest Diez Benavente, Roper C, Marinho CRF, Sutherland CJ, Hibberd ML, David Baker, Taane Clark, Susana Campino
External collaborators: Claudio Marinho (Univ. Sao Paulo, Brazil)
Funding Body: MRC
Malaria infection during pregnancy, caused by Plasmodium falciparum parasites, is a major public health burden in tropical areas of Africa and South East Asia, being responsible for substantial maternal and infant morbidity and mortality. The interaction between infected erythrocytes and the placental receptor is mediated by the parasite protein VAR2CSA. We aim to describe the genetic diversity of var2csa to help vaccine development.
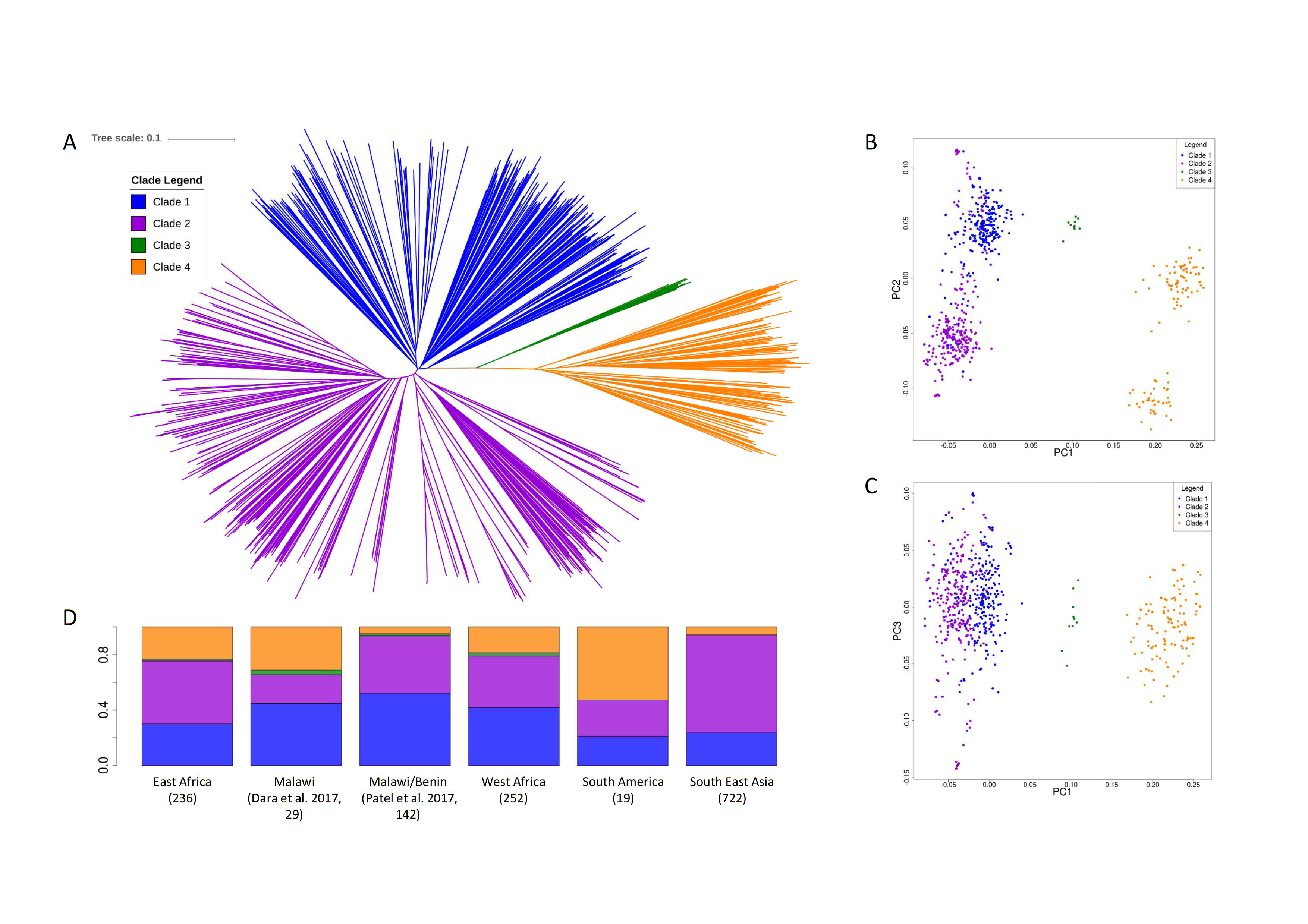
Malaria infection during pregnancy, caused by the sequestering of Plasmodium falciparum parasites in the placenta, leads to high infant mortality and maternal morbidity. The parasite-placenta adherence mechanism is mediated by the VAR2CSA protein, a target for natural occurring immunity. Currently, vaccine development is based on its ID1-DBL2Xb domain however little is known about the global genetic diversity of the encoding var2csa gene, which could influence vaccine efficacy. In a comprehensive analysis of the var2csa gene in >2,000P. falciparum field isolates across 23 countries, we found that var2csa is duplicated in high prevalence (>25%), African and Oceanian populations harbour a much higher diversity than other regions, and that insertions/ deletions are abundant leading to an underestimation of the diversity of the locus. Further, ID1-DBL2Xb haplotypes associated with adverse birth outcomes are present globally, and Africanspecific haplotypes exist, which should be incorporated into vaccine design.